For more information please visit the NI4OS-Europe service catalogue.

- Trainer: Enis Kocan
- Trainer: Zarko Zecevic
- Trainer: Žarko Zečević
For more information please visit the NI4OS-Europe service catalogue.

- Trainer: Enis Kocan
- Trainer: Zarko Zecevic
- Trainer: Žarko Zečević
For more information please visit the NI4OS-Europe service catalogue.

- Trainer: Tomislav Smuc
Training materials for the Air Pollution Prediction service.
The service is a simulation system for generation of prediction of air pollution levels based on WRF-Chem software. The outputs are hourly levels of air pollution for PM10, PM2.5, NO and SO2.
For more information please visit the NI4OS-Europe service catalogue.

- Trainer: Boro Jakimovski
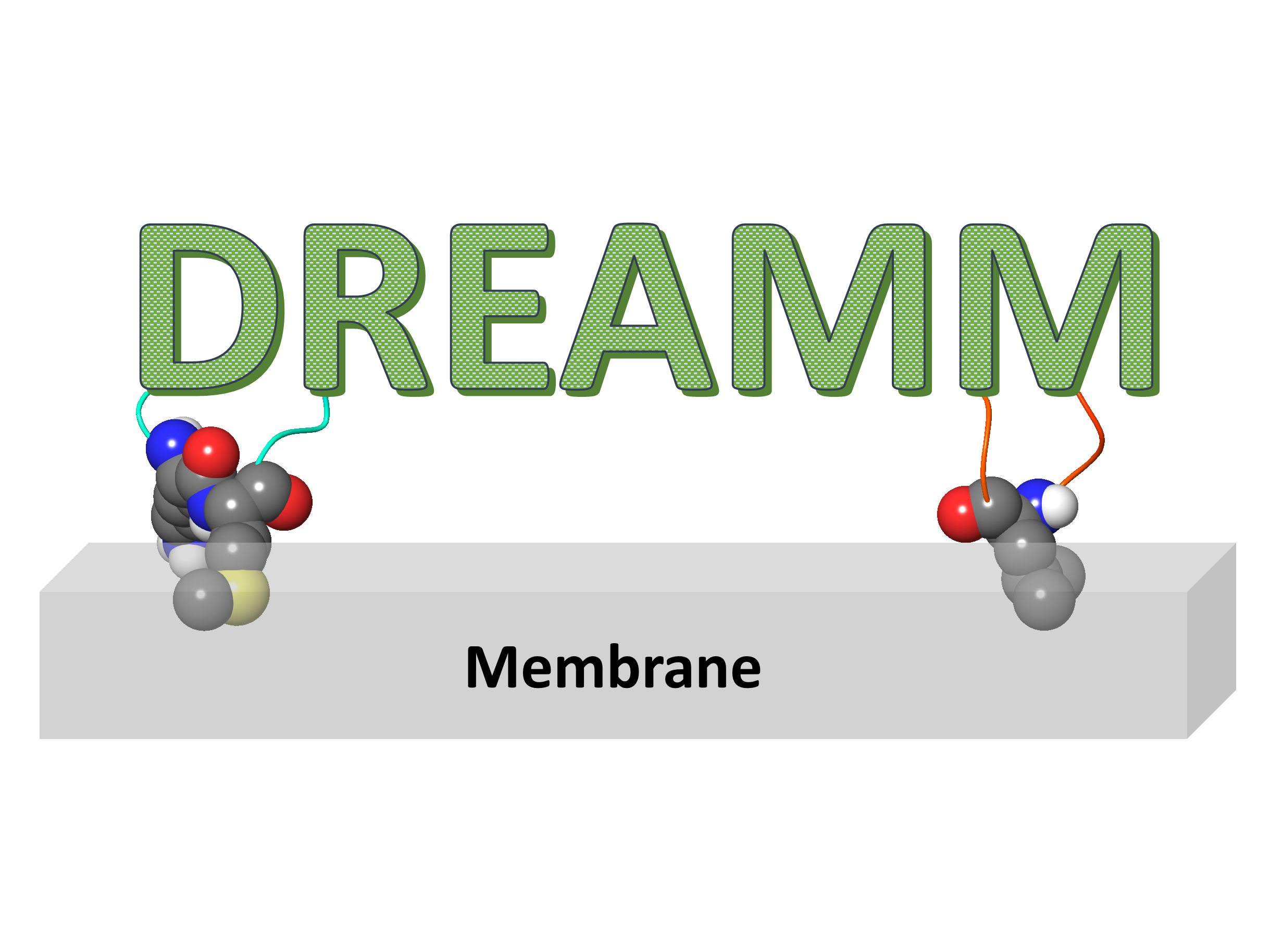
Training materials that help you learn what is and how to use the DREAMM service.
DREAMM is a novel machine learning tool that predicts the protein-membrane interfaces of peripheral membrane protein, and optionally predicts binding sites near the predicted membrane-penetrating residues in protein conformational ensembles. As an output, the user can retrieve the membrane-penetrating residues in a .csv file and if the user's choice was to predict binding sites, a .zip file will be downloaded including the above mentioned .csv file, the binding pocket predictions, the visualizations, and summarized the binding site clustering results.
More information about the service can be found in the NI4OS Service Catalogue.
After completing this course, you will:
- understand the benefits of the service,
- be familiar with the main functions of the service.
- Trainer: Zoe Cournia
Training materials that help you learn what is and how to use the ChemBioServer service.
ChemBioServer is a web-server for filtering, clustering and networking of chemical compound libraries facilitating both drug discovery and repurposing. It provides researchers the ability to (i) browse and visualize compounds along with their physicochemical and toxicity properties, (ii) perform property-based filtering of chemical compounds, (iii) explore compound libraries for lead optimization based on perfect match substructure search, (iv) re-rank virtual screening results to achieve selectivity for a protein of interest against different protein members of the same family, selecting only those compounds that score high for the protein of interest, (v) perform clustering among the compounds based on their physicochemical properties providing representative compounds for each cluster, (vi) construct and visualize a structural similarity network of compounds providing a set of network analysis metrics, (vii) combine a given set of compounds with a reference set of compounds into a single structural similarity network providing the opportunity to infer drug repurposing due to transitivity, (viii) remove compounds from a network based on their similarity with unwanted substances (e.g. failed drugs) and (ix) build custom compound mining pipelines.

- Trainer: Zoe Cournia
Training materials that help you learn what is and how to use the FEPrepare service.
FEP prepare is a webserver, which automates the set-up procedure for performing NAMD/FEP simulations. Automating free energy perturbation calculations is a step forward to delivering high throughput calculations for accurate predictions of relative binding affinities before a compound is synthesized, and consequently save enormous time and cost.

- Trainer: Zoe Cournia
Ingredio application is a natural processing language (NLP) application that offers a pipeline of three services related to biomedical text. The application is able to classify biomedical text based on certain features of its content, extract compound names and infer causal relations from the text, however it is experimental and is not meant to replace human curation. It's main use is to showcase how this can be used as a high-throughput and high precision language filtering software for large scale biomedical data.
For more information please visit the NI4OS-Europe catalogue.

- Trainer: Zoe Cournia
Self-paced course that introduces you to the Clowder flexible and extensible online content management system for Digital Cultural Heritage.
For more details on the service please visit the NI4OS-Europe service catalogue.

- Trainer: George Artopoulos
- Trainer: Georgios Artopoulos
Training materials on how to use the Gaussian API service from the field of Computational Physics. This is a service for fitting repulsive potentials in density-functional tight-binding with Gaussian process regression.
For more details on the service please visit the NI4OS-Europe service catalogue.

- Trainer: Bojana Koteska
Self-paced course that introduces you to the Live Access Server used for flexible access and visualization of geo-referenced scientific data.
For more details on the service please visit the NI4OS-Europe service catalogue.

- Trainer: Theodoros Christoudias
Training materials on how to use the Schrödinger thematic service used in Computational Physics. This is a service for advanced methods for solving of multidimensional time-independent Schrödinger equation.
For more information about the service please visit the NI4OS-Europe service catalogue.
After completing the course, you will:
- understand the scientific background of the service;
- gain the basic familiarity with Jupyter Notebooks;
- be familiar with the characteristics of the service,
- be familiar with the common workflows.

- Trainer: Bojana Koteska
Self-paced course that introduces you to NanoCrystal.
NanoCrystal is a novel web-based crystallographic tool that creates nanoparticle coordinates from any material crystal structure.
For more information please visit the NI4OS-Europe catalogue.

- Trainer: Zoe Cournia